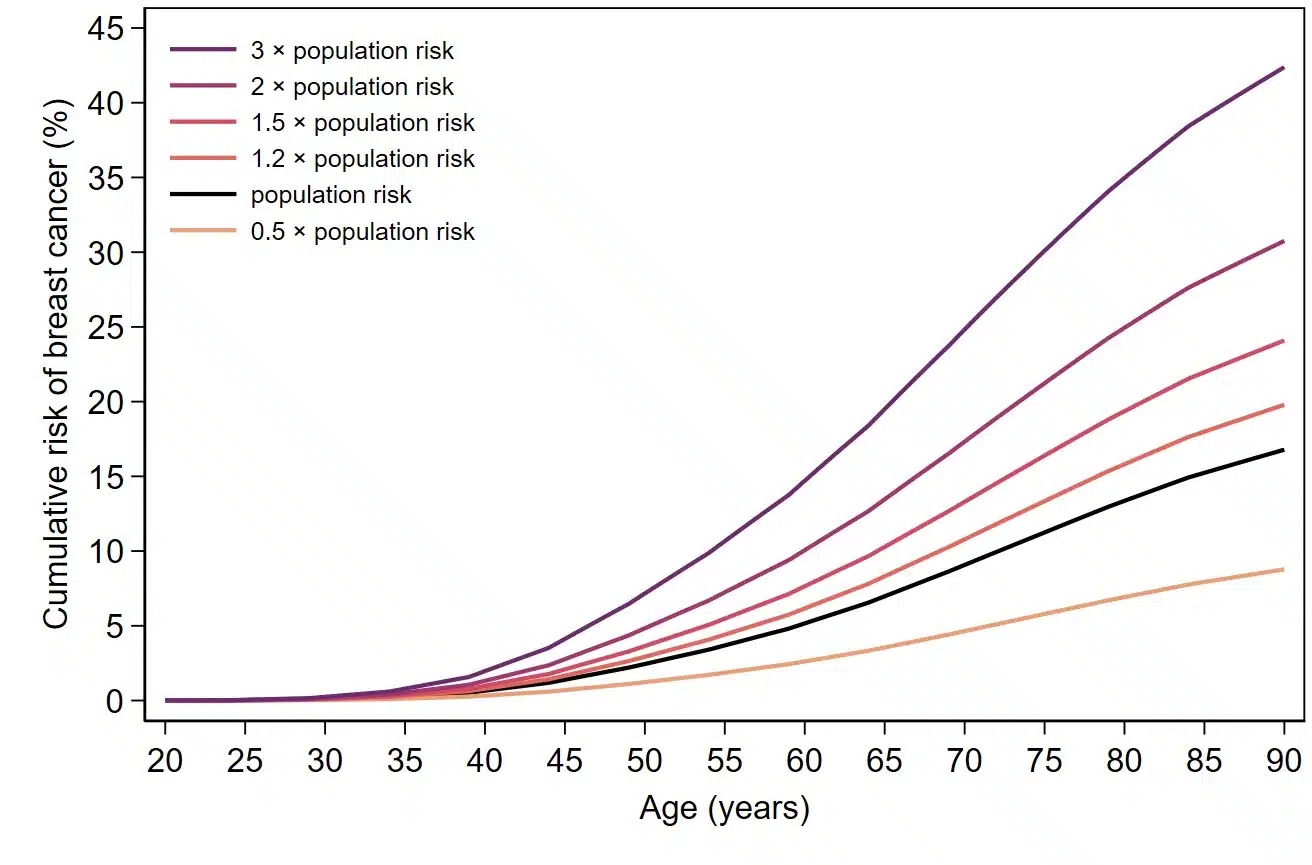
How does geneType™ measure up?
All geneType™ risk assessments are specifically developed, calibrated and validated for the general population. Direct comparisons against gold-standard clinical models are carried out in cohorts and/or case-control datasets using a variety of statistical methods common in the field of epidemiology.